Table of Contents
Auryn Python tools
Auryn v0.8.0 is the first version to come with a set of Python tools which allow decoding from binary files generated with BinarySpikeMonitor or BinaryStateMonitor. You can find the Python code in the tools/python directory.
To use the Auryn Python tools, point your Python path to the auryn/tools/python directory. For instance by
export PYTHONPATH=$PYTHONPATH:<path_to_auryn_src_directory>/tools/python
Example: Plot spikes from spk file
Suppose you have an spk file which was written by a BinarySpikeMonitor. For instance, if you run the example sim_coba_binmon this will by default write the file /tmp/coba.0.e.spk with spikes from the Vogels Abbott benchmark network.
The following code snipped will then plot the spikes from last 0.1 seconds in the file:
import numpy as np import pylab as pl from auryntools import * filename = "/tmp/coba.0.e.spk" seconds = 0.1 sf = AurynBinarySpikeFile(filename) spikes = np.array(sf.get_last(seconds)) pl.scatter(spikes[:,0], spikes[:,1]) pl.xlabel("Time [s]") pl.ylabel("Neuron ID") pl.show()
Instead of the the get_last() method you could have also use the get_spikes() method to get all or a temporal range of spikes from the file.
Example: Plot spikes from multiple spk files
In a parallel simulation you will typically have multiple spk output files because each rank writes its own file to disk. The Python toolkit provides a simple way how you can deal with this transparently. Suppose you ran the Vogels Abbott benchmark in parallel using 4 cores (mpirun -n 4 ./sim_coba_binmon). The following code will lets you plot the spikes:
import numpy as np import pylab as pl from auryntools import * num_mpi_ranks = 4 seconds = 0.1 filenames = [ "/tmp/coba.%i.e.spk"%i for i in range(num_mpi_ranks) ] sf = AurynBinarySpikeView(filenames) spikes = np.array(sf.get_last(seconds)) pl.scatter(spikes[:,0], spikes[:,1]) pl.xlabel("Time [s]") pl.ylabel("Neuron ID") pl.show()
The output should look similar to the plot above.
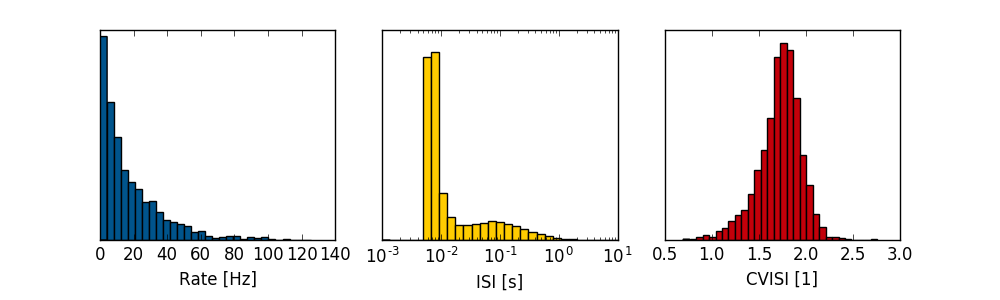
Example: Vogels plot (overview spiking statistics)
import numpy as np import pylab as pl from auryntools import * from auryntools.stats import * filename = "/tmp/coba.0.e.spk" sf = AurynBinarySpikeFile(filename) spikes = sf.get_spikes() vogels_plot(spikes)
Gives the following output on the above spiking data from the Vogels Abbott benchmark.
Example: Compute a neurons receptive field via reverse correlations
Suppose you want to know the linear receptive field of a neuron. We will illustrate this on data from one of my published papers http://www.nature.com/ncomms/2015/150421/ncomms7922/full/ncomms7922.html ( you can find the simulation code here https://github.com/fzenke/pub2015orchestrated). When you run this simulation with BinarySpikeMonitors (for instance the development branch of the above repository has that enabled by default), you will end up with multiple spk files for the input and the recurrent network dynamics. Here I will assume that you have done that already and the output files are accessible under the datapath which in the example is set to my home directory, but should be different in your case.
To get the receptive field of a neuron we are interested in which input neurons were active just before a certain network neuron spiked. Moreover, because the network is plastic we would like to know how the receptive field changes over time. With the supplied Python toolkit this analysis is straight forward. Here is the code:
#!/usr/bin/python import numpy as np import pylab as pl from auryntools import * datadir = "/home/zenke/data/sim" # Set this to your local data path num_mpi_ranks = 4 dim = 64 n_max = dim**2 t_bin = 100e-3 integration_time = 400 neuron_id = 28 outputfile = "%s/rf2.0.e.spk"%datadir sf = AurynBinarySpikeFile(outputfile) stimfiles = ["%s/rf2.%i.s.spk"%(datadir,i) for i in range(num_mpi_ranks)] sfo = AurynBinarySpikeView(stimfiles) start_times = np.arange(6)*500 for i,t_start in enumerate(start_times): t_end = t_start+integration_time print("Analyzing %is..%is"%(t_start,t_end)) spike_times = np.array(sf.get_spike_times(neuron_id, t_start, t_end)) hist = sfo.time_triggered_histogram( spike_times, time_offset=-t_bin, time_window=t_bin, max_neuron_id=n_max ) pl.subplot(2,3,i+1) pl.title("t=%is"%t_start) pl.imshow(hist.reshape((dim,dim)), origin='bottom') pl.show()
which will give you the following plot:
 as you can see the neuron here is torn between being selective to a square and a triangle.
as you can see the neuron here is torn between being selective to a square and a triangle.
Example: Get membrane potential from BinaryStateMonitor file
Finally, here is an example of howto read binary state monitor files:
import numpy as np import pylab as pl from auryntools import * # This code snipped assumes that you have run the example simulation # sim_epsp_binmon with default parameters and adjusted the below # filename to its output. filename = "../../build/release/examples/out_epsp.0.bmem" t_from=0.2 t_to =2.5 sf = AurynBinaryStateFile(filename) mem = np.array(sf.get_data(t_from, t_to)) pl.plot(mem[:,0], mem[:,1]) pl.xlabel("Time [s]") pl.ylabel("Membrane potential [V]") pl.show()

