This is an old revision of the document!
Table of Contents
Orchestrated Plasticity
You will find the code to reproduce the key results of Zenke, F., Agnes, E.J., and Gerstner, W. (2015). Diverse synaptic plasticity mechanisms orchestrated to form and retrieve memories in spiking neural networks. Nat Commun 6. on https://github.com/fzenke/pubsim
Getting the source
To run the orchestrated plasticity simulation as described in Figure 3 of the paper download or clone the pubsim repository. Then cd into /2015orchestrated/. You need to have Auryn installed and compiled. For this code to run you either need Auryn v0.5 or if you are used to working with Auryn v0.7 or newer, use the develop branch instead which supports Auryn name spaces (introduced with version v0.7).
Running the code
Now go to the installation directory of the simulation code (when you are
reading this, chances are you are already in this directory) and update the path to your auryn installation in Makefile. Then run make.
This should build the necessary Auryn libraries that implement plasticity
and the simulation libraries (*Connection.* and sim_*.cpp). For instance the binary file sim_rc_p10c is the
one behind Figure 3 in the paper.
Finally update the output path in globalvars.sh to point to a portion of disk
with sufficient space. If you want to run distributed simulations make sure this
path is accessible from all nodes of your cluster.
Invoking ./run_orchestrated_stdp.sh on the command line
will run the three scripts 1run_init.sh, 2run_learn.sh and 3run_cued.sh in
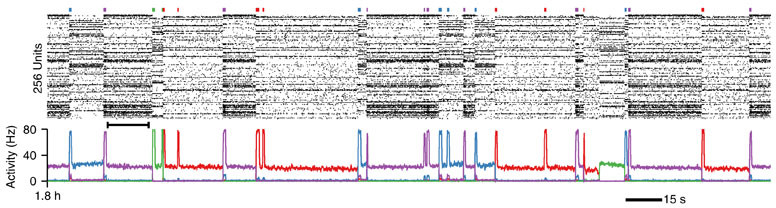
sequence. This corresponds to the first hours corresponding to the two-fold learning and recall protocol shown in Figure 3d and j.
Output files
The simulation will per default run on 4 cores locally, but this might change depending on your MPIsetup. It will generate a range of files out of which the most important ones are *.ras (or bras files if you use the newer develop branch version – to decode the “binary” ras files use aube). The ras files store the spike raster data of the simulation. Files labeled *.stimtimes contain the time point and duration of external stimulation events. These are text files. pact files contain the “Pattern Activity” in which patterns correspond to the cell assemblies that are formed during the course of the simulation. There is more information being saved, but these files will already allow you to reproduce Figure 3d and j (as shown above).
Updating rf1.pat to sort cells according to their preferred stimulus
Differences in the random seed or random library implementation might require to rebuild the rf1.pat file which stores the assembly membership of the individual neurons in the network. The file is used by PatternMonitor to read out the population firing rates of the four corresponding groups. To rebuild rf1.pat run the bootstrap.sh script. Note, that the script runs by default when you invoke ./run_orchestrated_stdp.sh.
That is also the reason why first phase of the simulation is run twice in case you wonder. After the first run the spiking data of the output is analyzed to determine cell/assembly membership relations by computing the PSTH for each cell and assigning cells to be selective to a certain pattern when their average firing rate during stimulation during the last bit of the simulation exceeds 10Hz. With the new rf1.pat generate the simulation can now rerun with the correct cells being recorded as the pattern activity (see Figure, below the spike raster).